| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2006-02-22 11:10:26 UTC |
|---|
| Update Date | 2023-02-21 17:15:51 UTC |
|---|
| HMDB ID | HMDB0001860 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Paraxanthine |
|---|
| Description | Paraxanthine, also known as p-xanthine, belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. Paraxanthine exists in all living organisms, ranging from bacteria to humans. Within humans, paraxanthine participates in a number of enzymatic reactions. In particular, paraxanthine and formaldehyde can be biosynthesized from caffeine; which is catalyzed by the enzyme cytochrome P450 1A2. In addition, paraxanthine and acetyl-CoA can be converted into 5-acetylamino-6-formylamino-3-methyluracil through its interaction with the enzyme arylamine N-acetyltransferase 2. In humans, paraxanthine is involved in caffeine metabolism. |
|---|
| Structure | CN1C=NC2=C1C(=O)N(C)C(=O)N2 InChI=1S/C7H8N4O2/c1-10-3-8-5-4(10)6(12)11(2)7(13)9-5/h3H,1-2H3,(H,9,13) |
|---|
| Synonyms | | Value | Source |
|---|
| 3,7-Dihydro-1,7-dimethyl-1H-purine-2,6-dione | ChEBI | | p-Xanthine | ChEBI | | 1,7-Dimethyl-xanthine | HMDB | | 1,7-Dimethylxanthine | HMDB, MeSH |
|
|---|
| Chemical Formula | C7H8N4O2 |
|---|
| Average Molecular Weight | 180.164 |
|---|
| Monoisotopic Molecular Weight | 180.06472552 |
|---|
| IUPAC Name | 1,7-dimethyl-2,3,6,7-tetrahydro-1H-purine-2,6-dione |
|---|
| Traditional Name | paraxanthine |
|---|
| CAS Registry Number | 611-59-6 |
|---|
| SMILES | CN1C=NC2=C1C(=O)N(C)C(=O)N2 |
|---|
| InChI Identifier | InChI=1S/C7H8N4O2/c1-10-3-8-5-4(10)6(12)11(2)7(13)9-5/h3H,1-2H3,(H,9,13) |
|---|
| InChI Key | QUNWUDVFRNGTCO-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | Xanthines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Xanthine
- 6-oxopurine
- Purinone
- Alkaloid or derivatives
- Pyrimidone
- Hydroxypyrimidine
- N-substituted imidazole
- Pyrimidine
- Imidazole
- Azole
- Heteroaromatic compound
- Lactam
- Azacycle
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | 351 - 352 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections |
|---|
| Predicted Molecular Properties | | Show more...
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| Paraxanthine,1TMS,isomer #1 | CN1C(=O)C2=C(N=CN2C)N([Si](C)(C)C)C1=O | 1875.3 | Semi standard non polar | 33892256 | | Paraxanthine,1TMS,isomer #1 | CN1C(=O)C2=C(N=CN2C)N([Si](C)(C)C)C1=O | 2088.9 | Standard non polar | 33892256 | | Paraxanthine,1TMS,isomer #1 | CN1C(=O)C2=C(N=CN2C)N([Si](C)(C)C)C1=O | 2612.9 | Standard polar | 33892256 | | Paraxanthine,1TBDMS,isomer #1 | CN1C(=O)C2=C(N=CN2C)N([Si](C)(C)C(C)(C)C)C1=O | 2095.9 | Semi standard non polar | 33892256 | | Paraxanthine,1TBDMS,isomer #1 | CN1C(=O)C2=C(N=CN2C)N([Si](C)(C)C(C)(C)C)C1=O | 2279.3 | Standard non polar | 33892256 | | Paraxanthine,1TBDMS,isomer #1 | CN1C(=O)C2=C(N=CN2C)N([Si](C)(C)C(C)(C)C)C1=O | 2605.4 | Standard polar | 33892256 |
| Show more...
|---|
| Spectra |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental GC-MS | GC-MS Spectrum - Paraxanthine GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-000i-2980000000-6ee5dc3100885f205536 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Paraxanthine GC-EI-TOF (Non-derivatized) | splash10-000i-2980000000-6ee5dc3100885f205536 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Paraxanthine GC-MS (Non-derivatized) - 70eV, Positive | splash10-0uka-3900000000-d979badf45de391835d4 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Paraxanthine GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine Quattro_QQQ 10V, Positive-QTOF (Annotated) | splash10-001i-0900000000-2d1aa5f80618f452686c | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine Quattro_QQQ 25V, Positive-QTOF (Annotated) | splash10-00di-0900000000-3326bcbee23b63dcc230 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine Quattro_QQQ 40V, Positive-QTOF (Annotated) | splash10-014l-9000000000-9824aef9dc2f13c24bcc | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive-QTOF | splash10-00e9-1900000000-9d0dde13bb1ce13e2644 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative-QTOF | splash10-00fr-0900000000-1aa733a1b7e41b994d8d | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine LC-ESI-qTof , Positive-QTOF | splash10-00di-1900000000-190ff3302e9f5a37a72f | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine LC-ESI-QTOF , negative-QTOF | splash10-00fr-0900000000-1aa733a1b7e41b994d8d | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine Linear Ion Trap , negative-QTOF | splash10-0229-0900000000-729a2bed81ba15260ffa | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine Linear Ion Trap , negative-QTOF | splash10-0229-0900000000-18d0a9c63a1dfb683c9b | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine , negative-QTOF | splash10-004i-0900000000-c52421f0ba1f46241853 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine LC-ESI-IT , positive-QTOF | splash10-00di-0900000000-0bd6ed3d764e5a9b374e | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine LC-ESI-QTOF , positive-QTOF | splash10-00e9-1900000000-9d0dde13bb1ce13e2644 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine Linear Ion Trap , positive-QTOF | splash10-0w29-0940000000-127f99d8e7324e01eb66 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine Linear Ion Trap , positive-QTOF | splash10-0ik9-0930000000-fe234438cbe3be0b30f6 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine , positive-QTOF | splash10-00di-1900000000-190ff3302e9f5a37a72f | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine , positive-QTOF | splash10-001i-0900000000-d6040ea84fe19d8c5cc6 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine 10V, Positive-QTOF | splash10-001i-0900000000-935e5a47dc1abe7b1196 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine 35V, Positive-QTOF | splash10-00di-3900000000-2f3998477ce19f3ff96f | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Paraxanthine 20V, Positive-QTOF | splash10-00di-4900000000-c89111b73e8a9a163111 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Paraxanthine 10V, Positive-QTOF | splash10-001i-0900000000-5e7c398698bb5c16617d | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Paraxanthine 20V, Positive-QTOF | splash10-00di-1900000000-522eaddc21ef92e70e62 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Paraxanthine 40V, Positive-QTOF | splash10-0005-9200000000-d10a1141c0e21f56ec00 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Paraxanthine 10V, Negative-QTOF | splash10-004i-1900000000-bc2d3cb520a4fcb81529 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Paraxanthine 20V, Negative-QTOF | splash10-004i-2900000000-e2e2992ce9b1265af790 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Paraxanthine 40V, Negative-QTOF | splash10-05am-9300000000-e1aefedceb8692ffe91f | 2017-09-01 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Experimental 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | 2012-12-04 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | 2012-12-05 | Wishart Lab | View Spectrum |
IR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M-H]-) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+H]+) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+Na]+) | 2023-02-03 | FELIX lab | View Spectrum |
| Show more...
|---|
| Biological Properties |
|---|
| Cellular Locations | - Cytoplasm (predicted from logP)
|
|---|
| Biospecimen Locations | - Blood
- Cerebrospinal Fluid (CSF)
- Feces
- Saliva
- Urine
|
|---|
| Tissue Locations | - Kidney
- Liver
- Placenta
- Prostate
|
|---|
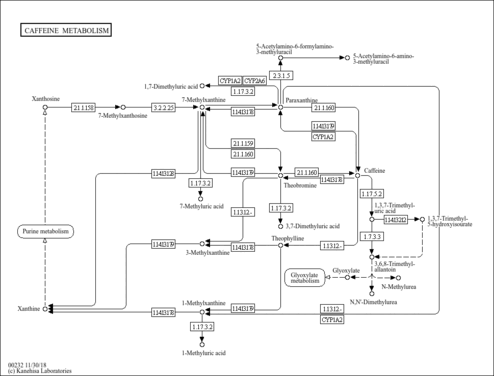
| Pathways | |
|---|
| Normal Concentrations |
|---|
| |
| Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 10.0 (0.30-28.0) uM | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Not Specified | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Urine | Detected and Quantified | 3.44 +/- 2 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 2.5 (3.2-4.0) umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details |
| Show more...
|---|
| Abnormal Concentrations |
|---|
| |
| Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal adenoma | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 1.780 +/- 0.641 uM | Adult (>18 years old) | Not Specified | Favorable outcome from traumatic brain injury | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 0.833 +/- 1.163 uM | Adult (>18 years old) | Male | Traumatic Brain Injury (TBI) | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 1.200 +/- 1.373 uM | Adult (>18 years old) | Female | Traumatic Brain Injury (TBI) | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 0.703 +/- 0.230 uM | Adult (>18 years old) | Not Specified | Traumatic brain injury | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 0.931 +/- 0.221 uM | Adult (>18 years old) | Not Specified | Traumatic brain injury (TBI) | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal Cancer | | details | | Urine | Detected and Quantified | 18.676 +/- 26.196 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Eosinophilic esophagitis | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal adenoma | | details | | Urine | Detected and Quantified | 3.6 (1.8-5.4) umol/mmol creatinine | Adult (>18 years old) | Both | Asthma | | details |
| Show more...
|---|
| Associated Disorders and Diseases |
|---|
| Disease References | | Traumatic brain injury |
|---|
- Sachse KT, Jackson EK, Wisniewski SR, Gillespie DG, Puccio AM, Clark RS, Dixon CE, Kochanek PM: Increases in cerebrospinal fluid caffeine concentration are associated with favorable outcome after severe traumatic brain injury in humans. J Cereb Blood Flow Metab. 2008 Feb;28(2):395-401. Epub 2007 Aug 8. [PubMed:17684518 ]
| | Head injury |
|---|
- Sachse KT, Jackson EK, Wisniewski SR, Gillespie DG, Puccio AM, Clark RS, Dixon CE, Kochanek PM: Increases in cerebrospinal fluid caffeine concentration are associated with favorable outcome after severe traumatic brain injury in humans. J Cereb Blood Flow Metab. 2008 Feb;28(2):395-401. Epub 2007 Aug 8. [PubMed:17684518 ]
| | Colorectal cancer |
|---|
- Brown DG, Rao S, Weir TL, O'Malia J, Bazan M, Brown RJ, Ryan EP: Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016 Jun 6;4:11. doi: 10.1186/s40170-016-0151-y. eCollection 2016. [PubMed:27275383 ]
- Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, Ahn J, Shi J, Sinha R: Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014 Sep;35(9):2089-96. doi: 10.1093/carcin/bgu131. Epub 2014 Jul 18. [PubMed:25037050 ]
| | Asthma |
|---|
- Zydron M, Baranowski J, Baranowska I: Separation, pre-concentration, and HPLC analysis of methylxanthines in urine samples. J Sep Sci. 2004 Oct;27(14):1166-72. [PubMed:15537072 ]
| | Eosinophilic esophagitis |
|---|
- Slae, M., Huynh, H., Wishart, D.S. (2014). Analysis of 30 normal pediatric urine samples via NMR spectroscopy (unpublished work). NA.
|
|
|---|
| Associated OMIM IDs | |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FooDB ID | FDB022714 |
|---|
| KNApSAcK ID | C00039930 |
|---|
| Chemspider ID | 4525 |
|---|
| KEGG Compound ID | C13747 |
|---|
| BioCyc ID | 1-7-DIMETHYLXANTHINE |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Paraxanthine |
|---|
| METLIN ID | 1457 |
|---|
| PubChem Compound | 4687 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 25858 |
|---|
| Food Biomarker Ontology | Not Available |
|---|
| VMH ID | Not Available |
|---|
| MarkerDB ID | Not Available |
|---|
| Good Scents ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Mueller, Christa E.; Deters, Dirk; Dominik, Andreas; Pawlowski, Maciej. Synthesis of paraxanthine and isoparaxanthine analogs (1,7- and 1,9-substituted xanthine derivatives). Synthesis (1998), (10), 1428-1436. |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| General References | - Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM: Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009 Feb 12;457(7231):910-4. doi: 10.1038/nature07762. [PubMed:19212411 ]
- Christensen M, Andersson K, Dalen P, Mirghani RA, Muirhead GJ, Nordmark A, Tybring G, Wahlberg A, Yasar U, Bertilsson L: The Karolinska cocktail for phenotyping of five human cytochrome P450 enzymes. Clin Pharmacol Ther. 2003 Jun;73(6):517-28. [PubMed:12811361 ]
- Fuhr U, Rost KL: Simple and reliable CYP1A2 phenotyping by the paraxanthine/caffeine ratio in plasma and in saliva. Pharmacogenetics. 1994 Jun;4(3):109-16. [PubMed:7920690 ]
- Tanaka E: Simultaneous determination of caffeine and its primary demethylated metabolites in human plasma by high-performance liquid chromatography. J Chromatogr. 1992 Mar 27;575(2):311-4. [PubMed:1629311 ]
- Horrigan LA, Kelly JP, Connor TJ: Immunomodulatory effects of caffeine: friend or foe? Pharmacol Ther. 2006 Sep;111(3):877-92. Epub 2006 Mar 15. [PubMed:16540173 ]
- Holstege A, Kurz M, Weinbeck M, Gerok W: Excretion of caffeine and its primary degradation products into bile. J Hepatol. 1993 Jan;17(1):67-73. [PubMed:8445222 ]
- Granfors MT, Backman JT, Laitila J, Neuvonen PJ: Oral contraceptives containing ethinyl estradiol and gestodene markedly increase plasma concentrations and effects of tizanidine by inhibiting cytochrome P450 1A2. Clin Pharmacol Ther. 2005 Oct;78(4):400-11. [PubMed:16198659 ]
- Blanchard J, Weber CW, Shearer LE: HPLC analysis of methylxanthines in human breast milk. J Chromatogr Sci. 1990 Dec;28(12):640-2. [PubMed:2292610 ]
- Delahunty T, Schoendorfer D: Caffeine demethylation monitoring using a transdermal sweat patch. J Anal Toxicol. 1998 Nov-Dec;22(7):596-600. [PubMed:9847011 ]
- Holland DT, Godfredsen KA, Page T, Connor JD: Simple high-performance liquid chromatography method for the simultaneous determination of serum caffeine and paraxanthine following rapid sample preparation. J Chromatogr B Biomed Sci Appl. 1998 Apr 10;707(1-2):105-10. [PubMed:9613939 ]
- Fuhr U, Rost KL, Engelhardt R, Sachs M, Liermann D, Belloc C, Beaune P, Janezic S, Grant D, Meyer UA, Staib AH: Evaluation of caffeine as a test drug for CYP1A2, NAT2 and CYP2E1 phenotyping in man by in vivo versus in vitro correlations. Pharmacogenetics. 1996 Apr;6(2):159-76. [PubMed:9156694 ]
- Zaigler M, Rietbrock S, Szymanski J, Dericks-Tan JS, Staib AH, Fuhr U: Variation of CYP1A2-dependent caffeine metabolism during menstrual cycle in healthy women. Int J Clin Pharmacol Ther. 2000 May;38(5):235-44. [PubMed:10839467 ]
- Blanchard J, Weber CW, Shearer LE: Methylxanthine levels in breast milk of lactating women of different ethnic and socioeconomic classes. Biopharm Drug Dispos. 1992 Apr;13(3):187-96. [PubMed:1576327 ]
- Wahllander A, Renner E, Karlaganis G: High-performance liquid chromatographic determination of dimethylxanthine metabolites of caffeine in human plasma. J Chromatogr. 1985 Mar 22;338(2):369-75. [PubMed:3998024 ]
- Koch JP, ten Tusscher GW, Koppe JG, Guchelaar HJ: Validation of a high-performance liquid chromatography assay for quantification of caffeine and paraxanthine in human serum in the context of CYP1A2 phenotyping. Biomed Chromatogr. 1999 Jun;13(4):309-14. [PubMed:10416066 ]
- Sachse C, Ruschen S, Dettling M, Schley J, Bauer S, Muller-Oerlinghausen B, Roots I, Brockmoller J: Flavin monooxygenase 3 (FMO3) polymorphism in a white population: allele frequencies, mutation linkage, and functional effects on clozapine and caffeine metabolism. Clin Pharmacol Ther. 1999 Oct;66(4):431-8. [PubMed:10546928 ]
- Elshenawy S, Pinney SE, Stuart T, Doulias PT, Zura G, Parry S, Elovitz MA, Bennett MJ, Bansal A, Strauss JF 3rd, Ischiropoulos H, Simmons RA: The Metabolomic Signature of the Placenta in Spontaneous Preterm Birth. Int J Mol Sci. 2020 Feb 4;21(3). pii: ijms21031043. doi: 10.3390/ijms21031043. [PubMed:32033212 ]
| Show more...
|---|