| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Expected but not Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2022-03-07 02:49:08 UTC |
|---|
| HMDB ID | HMDB0001175 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Malonyl-CoA |
|---|
| Description | Malonyl-CoA belongs to the class of organic compounds known as acyl-CoAs. These are organic compounds containing a coenzyme A substructure linked to an acyl chain. Thus, malonyl-CoA is considered to be a fatty ester lipid molecule. Malonyl-CoA is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. Within humans, malonyl-CoA participates in a number of enzymatic reactions. In particular, malonyl-CoA can be biosynthesized from acetyl-CoA; which is mediated by the enzyme acetyl-CoA carboxylase 1. In addition, malonyl-CoA can be converted into malonic acid and coenzyme A; which is catalyzed by the enzyme fatty acid synthase. Outside of the human body, malonyl-CoA has been detected, but not quantified in, several different foods, such as rapes, mamey sapotes, jew's ears, pepper (C. chinense), and Alaska wild rhubarbs. This could make malonyl-CoA a potential biomarker for the consumption of these foods. Malonyl-CoA is a coenzyme A derivative that plays a key role in fatty acid synthesis in the cytoplasmic and microsomal systems. |
|---|
| Structure | CC(C)(COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N)[C@@H](O)C(=O)NCCC(=O)NCCSC(=O)CC(O)=O InChI=1S/C24H38N7O19P3S/c1-24(2,19(37)22(38)27-4-3-13(32)26-5-6-54-15(35)7-14(33)34)9-47-53(44,45)50-52(42,43)46-8-12-18(49-51(39,40)41)17(36)23(48-12)31-11-30-16-20(25)28-10-29-21(16)31/h10-12,17-19,23,36-37H,3-9H2,1-2H3,(H,26,32)(H,27,38)(H,33,34)(H,42,43)(H,44,45)(H2,25,28,29)(H2,39,40,41)/t12-,17-,18-,19+,23-/m1/s1 |
|---|
| Synonyms | | Value | Source |
|---|
| coenzyme A, S-(Hydrogen propanedioate) | ChEBI | | Malonyl coenzyme A | ChEBI | | S-(Hydrogen malonyl)coenzyme A | ChEBI | | coenzyme A, S-(Hydrogen propanedioic acid) | Generator | | CoA, Malonyl | MeSH | | coenzyme A, Malonyl | MeSH | | Malonyl CoA | MeSH |
|
|---|
| Chemical Formula | C24H38N7O19P3S |
|---|
| Average Molecular Weight | 853.58 |
|---|
| Monoisotopic Molecular Weight | 853.115602295 |
|---|
| IUPAC Name | 3-[(2-{3-[(2R)-3-[({[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)methyl]-2-hydroxy-3-methylbutanamido]propanamido}ethyl)sulfanyl]-3-oxopropanoic acid |
|---|
| Traditional Name | malonyl-coa |
|---|
| CAS Registry Number | 524-14-1 |
|---|
| SMILES | CC(C)(COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N)[C@@H](O)C(=O)NCCC(=O)NCCSC(=O)CC(O)=O |
|---|
| InChI Identifier | InChI=1S/C24H38N7O19P3S/c1-24(2,19(37)22(38)27-4-3-13(32)26-5-6-54-15(35)7-14(33)34)9-47-53(44,45)50-52(42,43)46-8-12-18(49-51(39,40)41)17(36)23(48-12)31-11-30-16-20(25)28-10-29-21(16)31/h10-12,17-19,23,36-37H,3-9H2,1-2H3,(H,26,32)(H,27,38)(H,33,34)(H,42,43)(H,44,45)(H2,25,28,29)(H2,39,40,41)/t12-,17-,18-,19+,23-/m1/s1 |
|---|
| InChI Key | LTYOQGRJFJAKNA-DVVLENMVSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as acyl coas. These are organic compounds containing a coenzyme A substructure linked to an acyl chain. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl thioesters |
|---|
| Direct Parent | Acyl CoAs |
|---|
| Alternative Parents | |
|---|
| Substituents | - Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Aminopyrimidine
- Alkyl phosphate
- 1,3-dicarbonyl compound
- Imidolactam
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Monosaccharide
- Pyrimidine
- Fatty amide
- Phosphoric acid ester
- Tetrahydrofuran
- Imidazole
- Heteroaromatic compound
- Azole
- Thiocarboxylic acid ester
- Carbothioic s-ester
- Secondary alcohol
- Amino acid
- Carboxamide group
- Amino acid or derivatives
- Secondary carboxylic acid amide
- Organoheterocyclic compound
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Azacycle
- Oxacycle
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic oxygen compound
- Primary amine
- Hydrocarbon derivative
- Carbonyl group
- Organosulfur compound
- Organopnictogen compound
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Alcohol
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections| Adduct Type | Data Source | CCS Value (Å2) | Reference |
|---|
| [M-H]- | Baker | 253.053 | 30932474 |
|
|---|
| Predicted Molecular Properties | | Show more...
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatized | Show more...
|---|
| Spectra |
|---|
| MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Malonyl-CoA 10V, Positive-QTOF | splash10-000i-1912000130-da8095652669b99c8349 | 2015-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Malonyl-CoA 20V, Positive-QTOF | splash10-000i-0913000000-c93bc8aa729d6712ed08 | 2015-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Malonyl-CoA 40V, Positive-QTOF | splash10-000i-2911000000-dee360a0ee5e683d782c | 2015-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Malonyl-CoA 10V, Negative-QTOF | splash10-001i-9830140570-8e78a24b9858f67c1579 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Malonyl-CoA 20V, Negative-QTOF | splash10-001i-5910010010-97fdf25aafbfdff7c908 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Malonyl-CoA 40V, Negative-QTOF | splash10-057i-6900100000-33483ed329c67d3c4cac | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Malonyl-CoA 10V, Positive-QTOF | splash10-0udr-0300000090-543ec182be67d5b3f4d3 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Malonyl-CoA 20V, Positive-QTOF | splash10-000i-0901000020-8959c74e984991e0a14a | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Malonyl-CoA 40V, Positive-QTOF | splash10-0002-1109000000-8d09d9721850ff0a7162 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Malonyl-CoA 10V, Negative-QTOF | splash10-0udi-0000000090-c2d1e16bb980e1c364d7 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Malonyl-CoA 20V, Negative-QTOF | splash10-0f8d-5100001790-e6e13e3646b9b7044305 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Malonyl-CoA 40V, Negative-QTOF | splash10-0fba-9101304800-02ee50cea77f7be47ec3 | 2021-09-24 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum |
| Show more...
|---|
| Biological Properties |
|---|
| Cellular Locations | - Extracellular
- Membrane
- Mitochondria
- Peroxisome
|
|---|
| Biospecimen Locations | Not Available |
|---|
| Tissue Locations | - Adipose Tissue
- Fibroblasts
- Liver
- Pancreas
- Skeletal Muscle
|
|---|
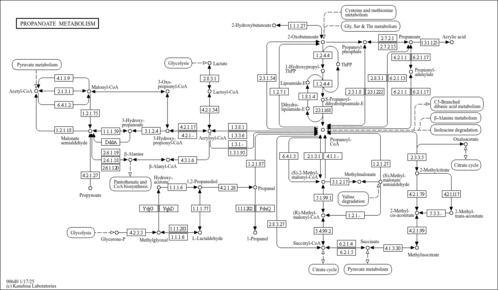
| Pathways | |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| Associated Disorders and Diseases |
|---|
| Disease References | None |
|---|
| Associated OMIM IDs | None |
|---|
| External Links |
|---|
| DrugBank ID | DB04524 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FooDB ID | FDB030990 |
|---|
| KNApSAcK ID | C00007260 |
|---|
| Chemspider ID | 559121 |
|---|
| KEGG Compound ID | C03188 |
|---|
| BioCyc ID | MALONYL-COA |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Malonyl-CoA |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 644066 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 15531 |
|---|
| Food Biomarker Ontology | Not Available |
|---|
| VMH ID | Not Available |
|---|
| MarkerDB ID | Not Available |
|---|
| Good Scents ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Hulsmann, W. C. Synthesis of malonyl coenzyme A from acetyl coenzyme A and oxalosuccinate in mitochondria. Biochimica et Biophysica Acta (1963), 77(3), 502-3. |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| General References | - Roepstorff C, Halberg N, Hillig T, Saha AK, Ruderman NB, Wojtaszewski JF, Richter EA, Kiens B: Malonyl-CoA and carnitine in regulation of fat oxidation in human skeletal muscle during exercise. Am J Physiol Endocrinol Metab. 2005 Jan;288(1):E133-42. Epub 2004 Sep 21. [PubMed:15383373 ]
- Napal L, Dai J, Treber M, Haro D, Marrero PF, Woldegiorgis G: A single amino acid change (substitution of the conserved Glu-590 with alanine) in the C-terminal domain of rat liver carnitine palmitoyltransferase I increases its malonyl-CoA sensitivity close to that observed with the muscle isoform of the enzyme. J Biol Chem. 2003 Sep 5;278(36):34084-9. Epub 2003 Jun 25. [PubMed:12826662 ]
- Kuhl JE, Ruderman NB, Musi N, Goodyear LJ, Patti ME, Crunkhorn S, Dronamraju D, Thorell A, Nygren J, Ljungkvist O, Degerblad M, Stahle A, Brismar TB, Andersen KL, Saha AK, Efendic S, Bavenholm PN: Exercise training decreases the concentration of malonyl-CoA and increases the expression and activity of malonyl-CoA decarboxylase in human muscle. Am J Physiol Endocrinol Metab. 2006 Jun;290(6):E1296-303. Epub 2006 Jan 24. [PubMed:16434556 ]
- Odland LM, Howlett RA, Heigenhauser GJ, Hultman E, Spriet LL: Skeletal muscle malonyl-CoA content at the onset of exercise at varying power outputs in humans. Am J Physiol. 1998 Jun;274(6 Pt 1):E1080-5. [PubMed:9611159 ]
- Bandyopadhyay GK, Yu JG, Ofrecio J, Olefsky JM: Increased malonyl-CoA levels in muscle from obese and type 2 diabetic subjects lead to decreased fatty acid oxidation and increased lipogenesis; thiazolidinedione treatment reverses these defects. Diabetes. 2006 Aug;55(8):2277-85. [PubMed:16873691 ]
- Pender C, Trentadue AR, Pories WJ, Dohm GL, Houmard JA, Youngren JF: Expression of genes regulating malonyl-CoA in human skeletal muscle. J Cell Biochem. 2006 Oct 15;99(3):860-7. [PubMed:16721829 ]
- Prentki M, Corkey BE: Are the beta-cell signaling molecules malonyl-CoA and cystolic long-chain acyl-CoA implicated in multiple tissue defects of obesity and NIDDM? Diabetes. 1996 Mar;45(3):273-83. [PubMed:8593930 ]
- Bavenholm PN, Pigon J, Saha AK, Ruderman NB, Efendic S: Fatty acid oxidation and the regulation of malonyl-CoA in human muscle. Diabetes. 2000 Jul;49(7):1078-83. [PubMed:10909961 ]
- Ruderman NB, Saha AK, Vavvas D, Witters LA: Malonyl-CoA, fuel sensing, and insulin resistance. Am J Physiol. 1999 Jan;276(1 Pt 1):E1-E18. [PubMed:9886945 ]
- Saha AK, Ruderman NB: Malonyl-CoA and AMP-activated protein kinase: an expanding partnership. Mol Cell Biochem. 2003 Nov;253(1-2):65-70. [PubMed:14619957 ]
- Morillas M, Gomez-Puertas P, Bentebibel A, Selles E, Casals N, Valencia A, Hegardt FG, Asins G, Serra D: Identification of conserved amino acid residues in rat liver carnitine palmitoyltransferase I critical for malonyl-CoA inhibition. Mutation of methionine 593 abolishes malonyl-CoA inhibition. J Biol Chem. 2003 Mar 14;278(11):9058-63. Epub 2002 Dec 23. [PubMed:12499375 ]
- Odland LM, Heigenhauser GJ, Lopaschuk GD, Spriet LL: Human skeletal muscle malonyl-CoA at rest and during prolonged submaximal exercise. Am J Physiol. 1996 Mar;270(3 Pt 1):E541-4. [PubMed:8638703 ]
- Trevisan CP, Angelini C, Fiorellini LA, Isaya G, Zacchello G: Malonyl-CoA abnormal inhibition of residual enzyme activity in carnitine palmitoyltransferase deficiency. Eur Neurol. 1986;25(4):309-16. [PubMed:3720808 ]
|
|---|