Summary
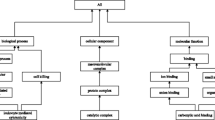
The Common Anatomy Reference Ontology (CARO) is being developed to facilitate interoperability between existing anatomy ontologies for different species, and will provide a template for building new anatomy ontologies. CARO has a structural axis of classification based on the top-level nodes of the Foundational Model of Anatomy. CARO will complement the developmental process sub-ontology of the GO Biological Process ontology, using the latter to ensure the coherent treatment of developmental stages, and to provide a common framework for the model organism communities to classify developmental structures. Definitions for the types and relationships are being generated by a consortium of investigators from diverse backgrounds to ensure applicability to all organisms. CARO will support the coordination of cross-species ontologies at all levels of anatomical granularity by crossreferencing types within the cell type ontology (CL) and the Gene Ontology (GO) Cellular Component ontology. A complete cross-species CARO could be utilized by other ontologies for cross-product generation.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
C. Berger, J. Urban, and G.M. Technau. Stage-specific inductive signals in the Drosophila neuroectoderm control the temporal sequence of neuroblast specification. Development, 128: 3243-3251, 2001.
J.A. Campos-Ortega and V. Hartenstein. TheEmbryonic Development of Drosophila melanogaster. (SecondEdition), Springer-Verlag, Berlin, 1999.
D.L. Cook, J.L.V. Mejino, and C. Rosse. Evolution of a foundational model of physiology: symbolic representation for functional bioinformatics.Medinfo, 11(1): 336-340, 2004.
D.H. Erwin and E.H. Davidson. The last common bilaterian ancestor. Development, 129:3021-3032, 2002.
Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res., 34: D322-6, 2006.
P.W.H. Holland. Major transitions in animal evolution: a developmental genetic perspective. American Zoologist, 38(6): 829-842,1998.
M. Jollie. Chordate Morphology. New York, Reinhold Books, 1962.
C.B. Kimmel, W.W. Ballard, S.R. Kimmel, B. Ullmann, and T.F. Schilling. Stages of embryonic development of the zebrafish.Dev. Dyn., 203: 253-310, 1995.
C. Mungall. Obol: integrating language and meaning in bio-ontologies. Comparative and Functional Genomics, 5 (6-7), 509-520, 2004.
P.D. Nieuwkoop and J. Faber. Normal Table of Xenopus laevis. 3rd Ed, 1994.
C. Rosse and J.L.V. Mejino. A reference ontology for bioinformatics: The Foundational Model of Anatomy. Journal of BiomedicalInformatics, 36: 478-500, 2003.
C. Rosse, J.L.V. Mejino, B.R. Modayur, R. Jakobovits, K.P. Hinshaw, J.F. Brinkley. Motivation and organizational principles for anatomicalknowledge representation: the Digital Anatomist symbolic knowledgebase. Journal of the American Medical InformaticsAssociation, 5(1): 17-40, 1998.
H. P. Schultze and M. Arsenault. The panderichthyid fish Elpistostege: A close relative of tetrapods? Paleontology, 28: 293-309, 1985.
M.A. Selleck and C.D. Stern. Fate mapping and cell lineage analysis of Hensen’s node in the chick embryo. Development, 112(2): 615-626, 1991.
J. Slack. From egg to Embryo (2nd Ed). Cambridge University Press, 1991.
B. Smith. Fiat objects. Topoi, 20(2): 131-148, 2001.
B. Smith, W. Ceusters, B. Klagges, J. KÖhler, A. Kumar, J. Lomax, C. Mungall, F. Neuhaus, A. Rector, and C. Rosse.Relations in biomedical ontologies. Genome Biology,6(5):r46, 2005.
B. Smith, W. Ceusters, and C. Rosse. On carcinomas and other pathological entities. Comparative and FunctionalGenomics, vol 6, 7-8, 379-387, 2005.
Editor information
Rights and permissions
Copyright information
© 2008 Albert Burger, Duncan Davidson, Richard Baldock
About this chapter
Cite this chapter
Haendel, M.A. et al. (2008). CARO – The Common Anatomy Reference Ontology. In: Burger, A., Davidson, D., Baldock, R. (eds) Anatomy Ontologies for Bioinformatics. Computational Biology, vol 6. Springer, London. https://doi.org/10.1007/978-1-84628-885-2_16
Download citation
DOI: https://doi.org/10.1007/978-1-84628-885-2_16
Publisher Name: Springer, London
Print ISBN: 978-1-84628-884-5
Online ISBN: 978-1-84628-885-2
eBook Packages: Computer ScienceComputer Science (R0)