Hammerhead
Rfam ID: RF00008
RF00163
RF02275
RF02276
RF02277
RF03152
click into different sections:
Timeline
-
1986 Discovery,Secondary structure of type I HHR[2]
-
1987 Secondary structure of type III HHR[3]
-
1991 Internal loops are important[4]
-
1994 Crystal structure of type III HHR[5]
-
1998 Dense positive charge is critical for catalysis[6]
-
2005 G8 and G12 is critical for catalysis[7]
-
2005 Catalytic mechanism[8]
-
2006 Crystal structure of type I HHR[9]
-
2011 Secondary structure of type II HHR[10]
-
2013 Profound catalytic mechanism[13]
-
2015 Trans-Hoogsteen greatly enhances the activity of the minimal hammer ribozyme[15]
-
2017 Found on some retrozyme sequences[17]
-
2017 Mg2+ is critical for catalysis by activating G12[18]
-
2018 Complex mechanism of enhancing activity[21]
Description
The hammerhead ribozyme is an RNA motif that catalyzes reversible cleavage and ligation reactions at a specific site within an RNA molecule. It is one of several catalytic RNAs (ribozymes) known to occur in nature. It serves as a model system for research on the structure and properties of RNA, and is used for targeted RNA cleavage experiments, some with proposed therapeutic applications. Named for the resemblance of early secondary structure diagrams to a hammerhead shark, hammerhead ribozymes were originally discovered in two classes of plant virus-like RNAs: satellite RNAs and viroids. They have subsequently been found to be widely dispersed within many forms of life.
Structure and mechanism
The secondary structure of the full-length hammerhead ribozyme with the general acid (red) and base (blue) is shown. The large arrow (baby blue) points to the scissile bond. The green line notes the tertiary interaction between the terminal loop of helix II and the internal loop in helix I.2D representation
|
|
3D visualisation
The overall structure of the hammerhead ribozyme is shown. This representation was generated from PDB ID: 3ZD5 at 2.2 Å resolution. Note the tertiary interaction between the terminal loop of helix II and the internal loop in helix I.
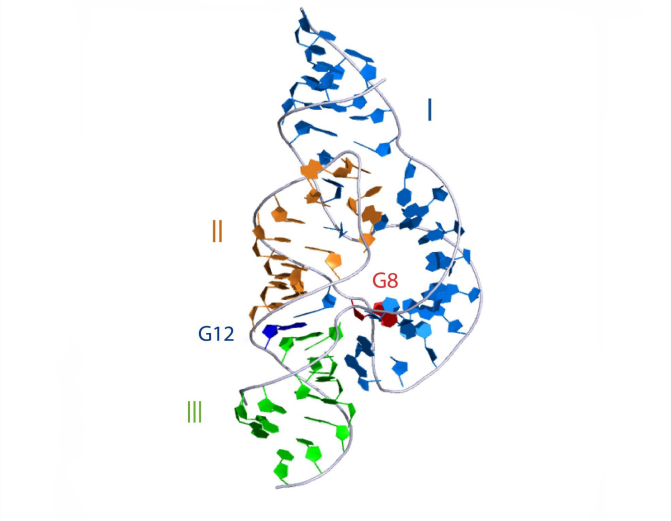 |
|
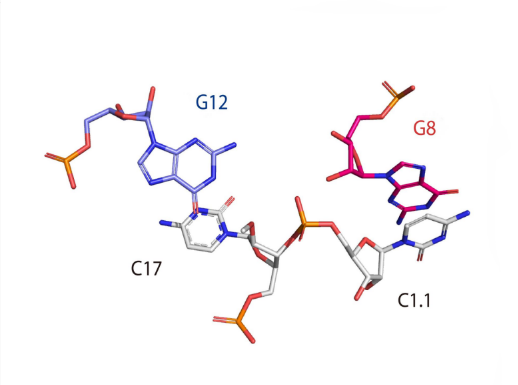
Catalytic centre
Close view of the catalytic center of the hammerhead ribozyme. This image was drawn using PDB ID: 3ZD5 at 2.2 Å resolution.
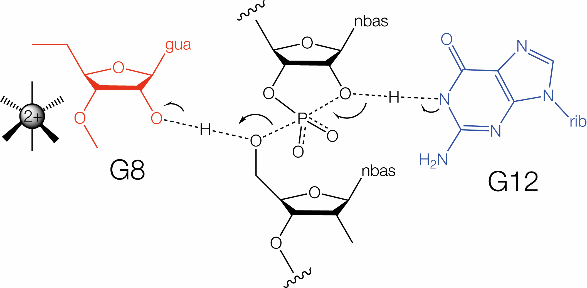
The hammerhead ribozyme mechanism is shown. G12 acts as the general base (blue) and G8 functions as the general acid (red).
 |
 |
Catalytic mechanism
The hammerhead ribozyme mechanism is reasonably well understood, although not totally established. The rate of cleavage rises across the whole accessible range of pH[24], suggesting that there are no participating groups of low pKa while substitution of G8 and G12 by 2,6-diaminopurine led to bell-shaped pH dependence[8], pointing to a role for these nucleotides in catalysis. In the crystal structure of the extended form of the hammerhead ribozyme the nucleobase of G12 is positioned with its N1 directed toward the O2' nucleophile. Its location thus suggests that the hammerhead is another ribozyme using N1 of a guanine nucleobase to act as the general base to remove the proton from the 2'-OH nucleophile in the cleavage reaction. This is supported by alkylation of G12 N1 using a substrate modified by 2'-bromoacetamide[25]. In the crystal the O2' of G8 is located adjacent to the O5' of the scissile phosphate, well positioned to donate a proton to the oxyanion leaving group. This is consistent with a marked alteration in the rate-pH profile for an atomic mutant where the 2' hydroxyl group of G8 was replaced by an amine[22]. The normal pKa of a 2'-hydroxyl group is 13.5. If it remains at this value in the hammerhead ribozyme, while most molecules will have a proton, it is very unlikely to be donated to another group. Therefore it seems probable that the pKa would need to be lowered, and the obvious way to achieve that would be juxtaposition of a positively-charged metal ion. QM/MM calculations suggested that a metal ion could move closer to lower the pKa of the G8 O2' in the transition state[26]. This is supported by the restoration of cleavage activity resulting either from the absence of divalent cations or the presence of a deoxyribose at G8 by 5'-phosphorothiolate substitution [25]. Moreover the cleavage rate exhibits a log-linear relationship as a function of the metal ion pKa [25]. In summary the hammerhead ribozyme appears to employ general acid-base catalysis using G N1 as the base, and a 2'-hydroxyl group (probably activated by a nearby metal ion) as general acid in cleavage. It is however conceivable that there is a second mechanistic channel whereby a water of hydration in the inner sphere of hydration of a bound metal ion acts as the general acid. This would then be closely similar to current ideas about the mechanism of the similar pistol ribozyme.
References
[1] Autolytic Processing of Dimeric Plant Virus Satellite RNA.
Prody, G. A., J. T. Bakos, J. M. Buzayan, I. R. Schneider and G. Bruening
Science 231(4745): 1577-1580.(1986)
[2] Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid.
Hutchins, C. J., P. D. Rathjen, A. C. Forster and R. H. Symons
Nucleic Acids Research 14(9): 3627-3640.(1986)
[3] Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites.
Forster, A. C. and R. H. Symons
Cell 49(2): 211-220.(1987)
[4] Newt satellite 2 transcripts self-cleave by using an extended hammerhead structure.
Pabón-Peña, L. M., Y. Zhang and L. M. Epstein
Mol Cell Biol 11(12): 6109-6115.(1991)
[5] Three-dimensional structure of a hammerhead ribozyme.
Pley, H. W., K. M. Flaherty and D. B. McKay
Nature 372(6501): 68-74.(1994)
[6] The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone.
Murray, J. B., A. A. Seyhan, N. G. Walter, J. M. Burke and W. G. Scott
Chem Biol 5(10): 587-595.(1998)
[7] Model for general acid-base catalysis by the hammerhead ribozyme: pH-activity relationships of G8 and G12 variants at the putative active site.
Han, J. and J. M. Burke
Biochemistry 44(21): 7864-7870.(2005)
[8] Model for general acid-base catalysis by the hammerhead ribozyme: pH-activity relationships of G8 and G12 variants at the putative active site.
Han, J. and J. M. Burke
Biochemistry 44(21): 7864-7870.(2005)
[9] Tertiary contacts distant from the active site prime a ribozyme for catalysis.
Martick, M. and W. G. Scott
Cell 126(2): 309-320.(2006)
[10] Capturing hammerhead ribozyme structures in action by modulating general base catalysis.
Chi, Y. I., M. Martick, M. Lares, R. Kim, W. G. Scott and S. H. Kim
PLoS Biol 6(9): e234.(2008)
[11] Structure-based search reveals hammerhead ribozymes in the human microbiome.
Jimenez, R. M., E. Delwart and A. Lupták
J Biol Chem 286(10): 7737-7743.(2011)
[12] Active-site monovalent cations revealed in a 1.55-Å-resolution hammerhead ribozyme structure.
Anderson, M., E. P. Schultz, M. Martick and W. G. Scott
J Mol Biol 425(20): 3790-3798.(2013)
[13] Bridging the gap between theory and experiment to derive a detailed understanding of hammerhead ribozyme catalysis.
Lee, T. S., K. Y. Wong, G. M. Giambasu and D. M. York
Prog Mol Biol Transl Sci 120: 25-91.(2013)
[14] Structural and catalytic effects of an invariant purine substitution in the hammerhead ribozyme: implications for the mechanism of acid-base catalysis.
Schultz, E. P., E. E. Vasquez and W. G. Scott
Acta Crystallogr D Biol Crystallogr 70(Pt 9): 2256-2263.(2014)
[15] Minimal Hammerhead Ribozymes with Uncompromised Catalytic Activity.
O'Rourke, S. M., W. Estell and W. G. Scott
J Mol Biol 427(14): 2340-2347.(2015)
[16] New classes of self-cleaving ribozymes revealed by comparative genomics analysis.
Weinberg, Z., P. B. Kim, T. H. Chen, S. Li, K. A. Harris, C. E. Lünse and R. R. Breaker
Nat Chem Biol 11(8): 606-610.(2015)
[17] Circular RNAs with hammerhead ribozymes encoded in eukaryotic genomes: The enemy at home.
de la Peña, M. and A. Cervera
RNA Biol 14(8): 985-991.(2017)
[18] Divalent Metal Ion Activation of a Guanine General Base in the Hammerhead Ribozyme: Insights from Molecular Simulations.
Chen, H., T. J. Giese, B. L. Golden and D. M. York
Biochemistry 56(24): 2985-2994.(2017)
[19] Numerous small hammerhead ribozyme variants associated with Penelope-like retrotransposons cleave RNA as dimers.
Lünse, C. E., Z. Weinberg and R. R. Breaker
RNA Biol 14(11): 1499-1507.(2017)
[20] Structure-based mechanistic insights into catalysis by small self-cleaving ribozymes.
Ren, A., R. Micura and D. J. Patel
Curr Opin Chem Biol 41: 71-83.(2017)
[21] Structural Simplicity and Mechanistic Complexity in the Hammerhead Ribozyme.
O'Rourke, S. M. and W. G. Scott
Prog Mol Biol Transl Sci 159: 177-202.(2018)
[22] Comparison of the Structures and Mechanisms of the Pistol and Hammerhead Ribozymes.
Wilson, T. J., Y. Liu, N. S. Li, Q. Dai, J. A. Piccirilli and D. M. J. Lilley
J Am Chem Soc 141(19): 7865-7875.(2019)
[23] Detection of Low-Abundance Metabolites in Live Cells Using an RNA Integrator.
You, M., J. L. Litke, R. Wu and S. R. Jaffrey
Cell Chem Biol 26(4): 471-481.e473.(2019)
[24] The hammerhead cleavage reaction in monovalent cations.
Curtis, E.A. and Bartel, D.P.
RNA, 7, 546-552.(2001)
[25] Probing general acid catalysis in the hammerhead ribozyme.
Thomas, J.M. and Perrin, D.M.
J. Amer. Chem. Soc., 131, 1135-1143.(2009)
[26] Role of Mg2+ in hammerhead ribozyme catalysis from molecular simulation.
Lee, T.S., Silva Lopez, C., Giambasu, G.M., Martick, M., Scott, W.G. and York, D.M.
J. Amer. Chem. Soc., 130, 3053-3064.(2008)
 Home
Home Database
Database Research
Research About us
About us



 3zd5
3zd5